Stanford Scientists Investigate TB Immune Response with Spatial Proteomics Technology
Research team aimed to identify and map immune players in tuberculosis granulomas
Researchers have long known that the immune response to the Mycobacterium tuberculosis pathogen leads to the formation of granulomas, or clusters of immune cells designed to barricade the bacteria and prevent the spread of infection in the lungs. But the inability to study these granulomas individually — and at the single-cell level — has stymied discovery and development of therapies for tuberculosis (TB).
For all the decades of research that have gone into TB, the disease remains a largely unsolved problem, says Erin McCaffrey, a graduate student focused on immunology in the Angelo Lab at Stanford University. Globally, TB leads to almost 1.5 million deaths each year.
It is also a seriously complex infection. Granulomas, which are just part of the infection cycle, have not yet been fully characterized. Research has suggested that granuloma structure is important in TB, and basic histological analyses have shown that certain immune cells may be restricted to specific areas of the granuloma. Until recently, these elements had not been mapped out at the cellular and molecular level in humans.
McCaffrey and her collaborators at Stanford and other institutions hypothesized that a more comprehensive, high-resolution view of granulomas throughout the infection cycle would reveal important biological mechanisms associated with TB infection and the immune response. Their goal was ambitious: identify all of the immune players involved in TB granulomas and map their spatial context to understand the interactions and complex organization of these elements. “The way cells are organized in the tissue is just as important as the types of cells that are present,” McCaffrey says.
What the team found has major implications for TB research and potentially could even improve the diagnosis of this infection. In a preprint posted to bioRxiv, the scientists report what they believe to be “the most comprehensive systems analysis of TB to date.”
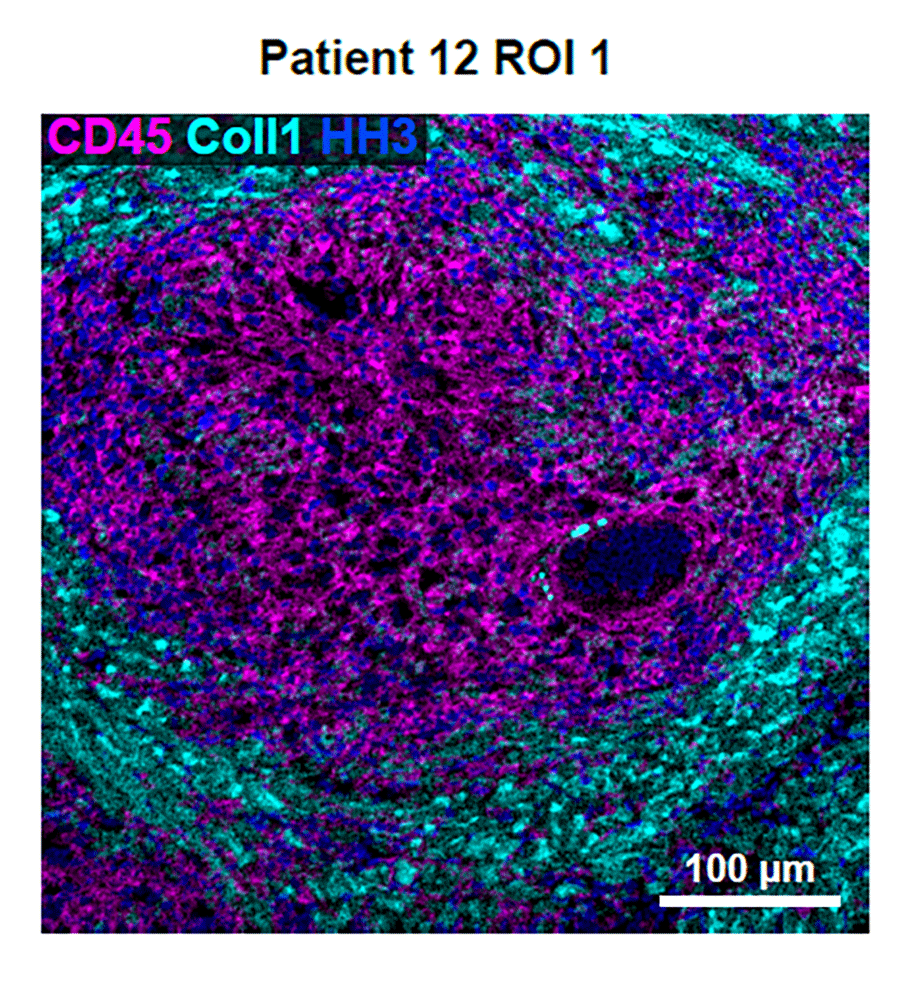
MIBI technology enabled a spatially resolved microenvironment profile
Fortunately, this team had access to a novel method that would make all the difference — multiplexed ion beam imaging (MIBI™). MIBI technology, originally developed at Stanford and commercialized by Ionpath, is a high-definition spatial proteomics platform that deploys the power of mass spec analysis to generate a highly multiplexed image and spatially resolved single-cell profile of the tissue microenvironment.
McCaffrey’s project involved MIBI imaging analysis of FFPE tissue samples collected from 13 patients with active tuberculosis infections. Regions in two granulomas from each patient were scanned. MIBI imaging enabled the team to simultaneously visualize and quantify dozens of biomarkers. The resulting spatial proteomic data powered analyses that led to insights about cellular heterogeneity, quantification of key proteins, and spatial context of individual single cells within the granuloma microenvironment.
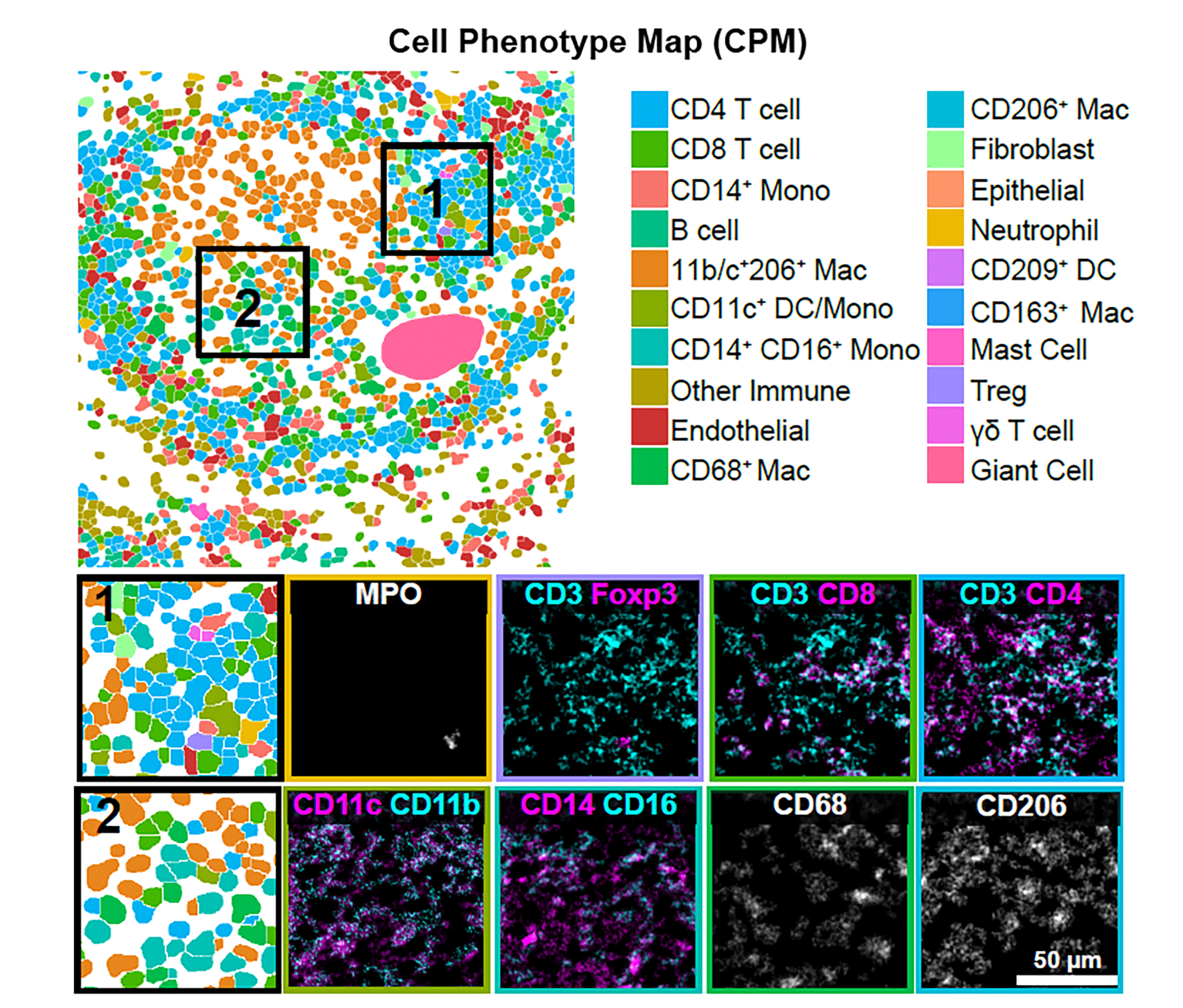
Spatial proteomic analysis revealed distinct phenotype profiles
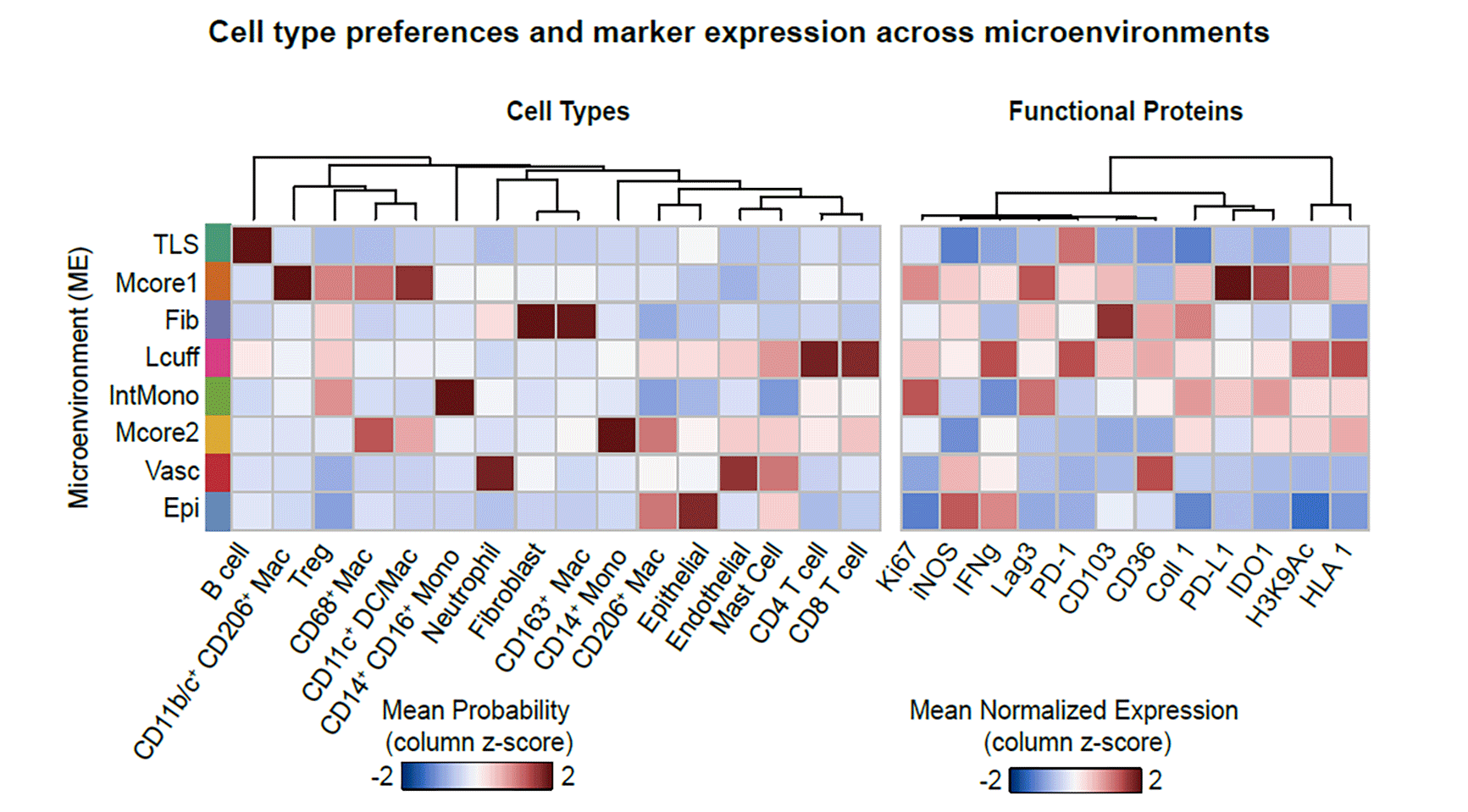
Based on that information, McCaffrey and her collaborators observed and profiled 19 distinct cell subsets and mapped them to eight phenotypically different granuloma microenvironments with varying roles in TB. For instance, they learned that the core of the granuloma is a highly immune-suppressed environment — similar to that seen in some tumors. Unlike the tumor microenvironment, T cells exhibited little evidence of activation, rather than being exhausted as expected. “This is a feature that was very consistent across a number of different samples we looked at,” says McCaffrey, who is lead author of the paper.
The preprint authors report that the microenvironment is characterized by “spatially coordinated co-expression of IDO1 and PD-L1 by myeloid cells and proliferating regulatory T cells.” While some features they found are common in the immune response to other conditions, the link between IDO1 and PD-L1 was not. “Even compared to sarcoidosis, foreign body uptake, xanthomatosis, and endometrial lesions, spatially coordinated expression of IDO1 and PD-L1 was unique to mycobacterial granulomas,” the scientists note.
By comparing their results to gene expression data produced in numerous studies encompassing more than 1,500 TB patients, the team also found that some of the same hallmarks they saw in granuloma biology mirrored patterns in patients’ blood. Where granulomas featured high expression of PD-L1, for instance, there were also high levels of PD-L1 in the blood, and they correlated with TB progression and response to treatment. “This is one of the most interesting aspects of the study,” McCaffrey adds. “It has us thinking about clinical biomarkers for TB.” PD-L1 gene expression may prove to be a straightforward blood-based marker for progression of the infection or treatment response.
The team also constructed a comprehensive TB granuloma tissue atlas with cell lineage, functional state, and spatial distribution information. “The work presented here reveals new aspects of immune regulation in TB infection that have important implications for understanding disease pathogenesis and improving clinical management,” McCaffrey et al. report.
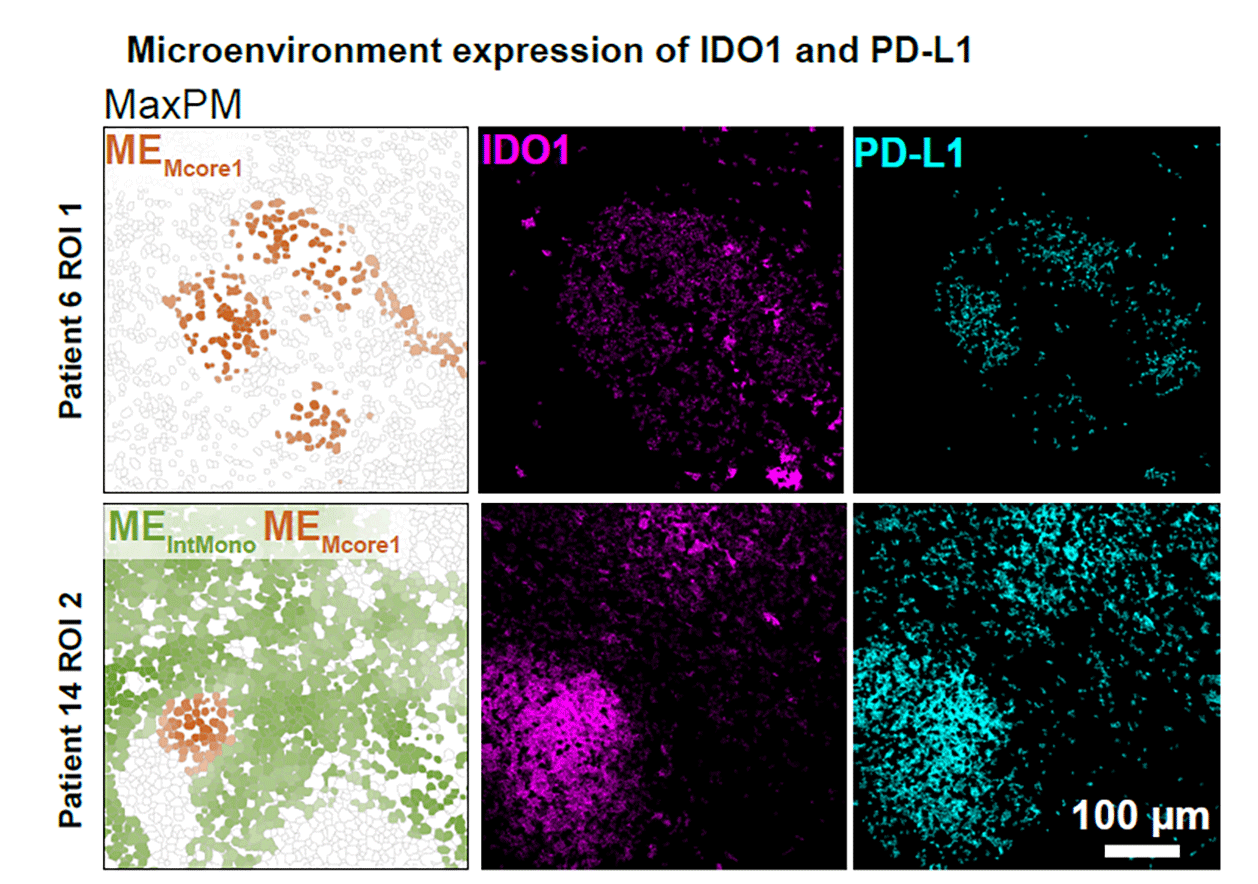
Why MIBI matters
“MIBI has advantages over other approaches,” McCaffrey says. One is the ability to simultaneously stain an image for all desired proteins, rather than the time-consuming serial method used by other spatial technologies. Another is its ability to process FFPE samples. TB is classified as a BSL-3 pathogen; any research with the organism is restricted to a small number of laboratories. “But in FFPE format it becomes safer to work with and transfer,” McCaffrey says. Since FFPE samples for TB patients were readily available, MIBI afforded the Stanford team a critical degree of flexibility for this research.
How future spatial biology projects will offer novel insights
Looking ahead, McCaffrey and her colleagues hope to perform follow-on studies that will continue to elucidate the immune response to TB. Functional models will be important, as will additional MIBI work. As the scientists write, “Future multiplexed imaging studies of granulomas from controlled TB exposures will offer novel insights on how these local regulatory dynamics influence granuloma fate, and ultimately, infection outcome.”
Beyond TB, McCaffrey hopes to see more researchers adopt spatial biology tools. “There are a lot of interesting ideas and new hypotheses you can glean when the spatial context is maintained,” she says. “The more people who collect these data sets with spatial information, the more we’ll learn.”